Jan 23, 2023 - Atlanta, GA
Georgia Tech researchers have recently published a unique paper that incorporates results from a teaching lab where undergraduates were given the opportunity to contribute a bulk of the research. The College of Sciences Dean Susan Lozier shares her thoughts on the project:
“So much of what we hope to achieve in the College is manifested in this paper: a collection of undergraduates with different majors have worked with several faculty members to produce publishable research that illustrates the arc from fundamental science to drug discovery. I cannot emphasize how impressed I am with this effort. Kudos to all involved.”

Undergraduate students performing bacterial isolation experiments in recently renovated laboratory space in the Boggs building.
“When the COVID pandemic hit, undergraduate researchers were the first to be excluded from research labs due to social distancing requirements. This had a serious negative impact on their training in experimental sciences. We are working to resolve that via pedagogic innovation and new curriculum design in laboratory coursework,” said the Drs. Vinayak Agarwal and Neha Garg who spearheaded this effort together with the Dr. Anton Bryksin who leads the Molecular Evolution Core in the Parker H. Petit Institute for Bioengineering and Bioscience.
Faculty involved believe that early training at the undergraduate level in the understanding and execution of these workflows will prepare and arm our STEM workforce to take on advanced challenges in modern-day research. This exposure to cutting-edge infrastructure employed by scientists helps them to better understand the functioning of life forms at an earlier level.
Lead author Jessica Deutsch and Dr. Neha Garg demonstrate mass spectrometry data collection.
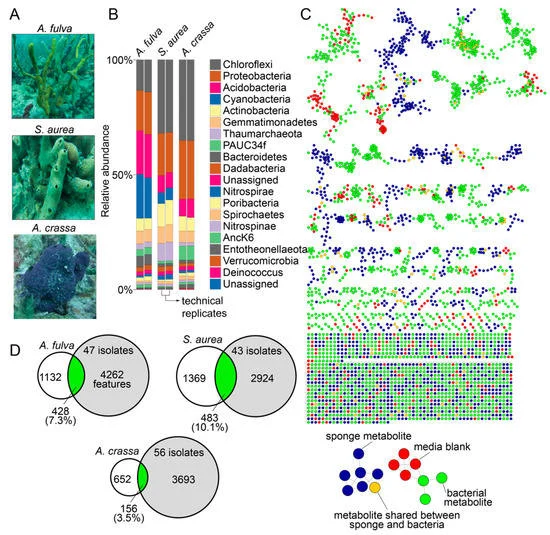
In this study, the undergraduates isolated bacteria from the marine environment, and generated DNA sequencing and mass spectrometry-based metabolomics data to find new-to-science small organic molecules called Natural Products. “Natural Products produced by the microbiome affect the composition of microbial community present in an organism,” Dr. Garg explains. “By doing so they affect the fitness of the organism in an environment and its resilience to pathogens and diseases. Access to advanced mass spectrometry instrumentation and machine learning methods to analyze large datasets allow linking of natural products to producing organisms. Thus, these research workflows are an integral part of delineating the role of microorganisms and their metabolic output in the fitness of an organism.”
“Undergraduates can, and often do, perform at the highest levels in original research,” says Dr. MG Finn, Chair of the chemistry department. “This paper provides an outstanding example on a problem of major importance to chemistry, drug discovery, and marine science. Congratulations to all involved!”
Link for published paper: Limited Metabolomic Overlap between Commensal Bacteria and Marine Sponge Holobionts Revealed by Large Scale Culturing and Mass Spectrometry-Based Metabolomics: An Undergraduate Laboratory Pedagogical Effort at Georgia Tech
